Epigenetic Diseases
Silver-Russell Syndrome (SRS) and Beckwith-Wiedemann Syndrome (BWS) are two severe disorders affecting fetal growth in humans. Children with SRS are typically short, with facial dysmorphia, macrocephaly, skeletal asymmetry, early puberty, and genital abnormalities. BWS is a complex disorder characterized by fetal overgrowth, enlarged internal organs, and developmental anomalies at birth, which increase the risk of embryonic tumors. In some cases, these syndromes have a genetic cause, but more often, they are due to an epigenetic alteration in genes subjected to genomic imprinting.
Imprinted genes are genes expressed from only one allele—either maternal or paternal—while the other is silent. About 70 imprinted genes have been identified in humans, most of which have a mouse counterpart that is also imprinted. These genes primarily influence embryonic and placental development, cell proliferation control, and behavior.
Imprinting Regulation
Imprinted genes are usually clustered in the human and mouse genomes. These large genetic areas, called imprinting domains, are regulated by imprinting control regions (ICRs). The maternal allele of an ICR carries different epigenetic marks from the paternal allele, with only one being active, thus regulating the gene in an allele-specific manner.
Robert Feil, Research Director at the Institute of Molecular Genetics in Montpellier, has been studying this subject for five years. “It’s typically DNA methylation that regulates ICRs by affecting one of the two copies. But other epigenetic modifications can also be involved in this control. We showed with David Umlauf and Katia Delaval that chemical modifications of histones, such as methylation or acetylation, can also determine imprinting” (Umlauf D et al., Nature Genet 2004; Delaval K, Feil R. Curr Opin Genet Dev 2004). During individual development, the imprinting of ICRs is maintained in somatic cells, but in germline cells, it is erased and reset during gametogenesis. “The mechanisms for maintaining imprinting in somatic cells are still largely unknown, but we know that DNA methyltransferases, the enzymes that methylate DNA, are involved in this process,” Feil adds. “We also showed that enzymes modifying histones are involved” (Fournier et al. EMBO J, 2002).
IGF2-H19 and KCNQ1 Domains
One of the multiple causes of Beckwith-Wiedemann Syndrome (BWS) is the overexpression of IGF2 (Insulin-like Growth Factor 2), a gene coding for a growth factor expressed only from the paternal allele and involved in fetal growth. IGF2 is located on human chromosome 11p15, which contains two imprinting domains: IGF2-H19 and KCNQ1. “A similar organization is found in mice on chromosome 7, making it a good model for studying the epigenetic mechanisms involved in SRS and BWS,” says Feil, who has long worked with the mouse model.
When IGF2 is overexpressed in mice, they exhibit an overgrowth phenotype from early fetal development. Conversely, when IGF2 expression is reduced, the mice are small. Next to IGF2 lies the H19 gene, which encodes a non-coding RNA of unknown function. The ICR of this domain is upstream of H19.
On the maternal allele, the ICR is unmethylated, allowing proteins to bind to this ICR, isolating the IGF2 gene from its downstream enhancer, preventing its transcription. H19 is transcribed on this maternal allele. On the paternal allele, however, the ICR is methylated, preventing proteins from binding to the DNA. Consequently, the enhancer is not isolated from IGF2, which is transcribed, while the paternal allele of H19 remains silenced.
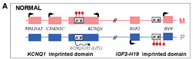
Epigenetic Etiology of SRS and BWS
The epigenetic marks on ICRs are set in germ cells and maintained during development post-fertilization. “If these epigenetic marks are not properly maintained during embryogenesis, the baby can develop diseases or an increased risk of certain cancers,” Feil notes.
Teams led by Yves Le Bouc, director of INSERM Unit 513 “Growth, Differentiation, and Tumor Processes” in Paris, identified a loss of methylation at the H19 ICR in one-third of SRS patients (Gicquel et al., Nat Genet, 2005). SRS typically occurs sporadically, with a normal karyotype in patients. When there is a loss of methylation on the paternal allele, IGF2 expression is significantly reduced, leading to dwarfism, while H19 is expressed from both alleles.
Conversely, in 10% of BWS cases, the syndrome is associated with a gain of methylation on the H19 ICR, resulting in biallelic expression of IGF2 and explaining the overgrowth, while H19 is silenced. BWS occurs sporadically in most cases after epigenetic changes in the 11p15 region.
In half of BWS cases, deregulation at the KvDMR1 ICR is responsible. Loss of methylation at this site induces biallelic expression of the non-coding RNA KCNQ1OT1 and reduced expression of CDKN1C, a growth-limiting factor, suggesting it may be responsible for the syndrome. Some BWS cases are indeed caused by a genetic mutation in CDKN1C, supporting this hypothesis.
Early Developmental Accident
But when do the epigenetic changes leading to BWS and SRS occur? It seems they happen early in development, after fertilization, according to Feil. “In most SRS patients, the DNA methylation loss at the H19 ICR does not affect all blood cells. Similarly, fibroblasts derived from tissues show mosaic methylation loss.” This observation suggests that the methylation loss did not occur in the sperm, as all cells would otherwise show a homogeneous loss of methylation.
For BWS, whether due to a loss of methylation at KvDMR1 or a gain at the H19 ICR, the epigenetic alteration likely occurs early in embryonic development. However, the possibility of it happening in the germline, during gamete formation, cannot be ruled out.
Studies on monozygotic twins strengthen the hypothesis of an early embryonic event. In identical twins originating from a single egg that splits, one twin can have SRS while the other remains healthy, indicating that the epigenetic transformation occurred after the embryo split. Similar observations have been made in BWS-affected twins, where one twin may have BWS due to KvDMR1 methylation loss while the other does not.
X Chromosomes Implicated
“In collaboration with Neil Brockdorff from London, we tested this hypothesis using XX undifferentiated mouse embryonic stem (ES) cells. In various XX ES cell lines where both X chromosomes are active, we observed numerous hypomethylated DNA sequences. We examined the methylation status of H19 and KCNQ1 imprinting control regions. The paternal H19 ICR was hypomethylated—normally methylated—while KvDMR1 was hypomethylated, usually methylated on the maternal allele. This hypomethylation was associated with decreased levels of DNA methyltransferases Dnmt3a and Dnmt3b. After in vitro differentiation, these cells did not regain their normal methylation status at these ICRs” (Zvetkova et al., Nature Genet, 2005).
Varied Phenotypes
Does the SRS phenotype vary depending on the timing of the methylation loss at the H19 ICR? If it occurs early, most or all cells are affected. If it occurs later, fewer cells are affected. Indeed, there is a correlation. In patients with the full SRS spectrum, complete methylation loss is observed in this region, whereas mosaic individuals have a milder syndrome.
A Maintenance Problem
If imprinting alterations responsible for BWS and SRS occur early in embryonic development, they were not present in the egg, indicating an issue in imprinting maintenance. This process, involving the upkeep of chromatin methylation and associated protein modifications, remains largely a mystery.
A similar issue arises in embryonic stem cells and embryos extracted from their natural environment and cultured in vitro. After IVF with intracytoplasmic sperm injection (ICSI), BWS and SRS frequencies increase, though the causal mechanism remains unknown. These manipulations may predispose to epigenetic instability in imprinting control regions.
Understanding allele-specific methylation maintenance at ICRs is essential for grasping the etiology of disorders like SRS and BWS. This research currently occupies part of Robert Feil’s team. “We are particularly interested in the role of histone methylation in this mechanism. Alexandre Wagschal, for example, is trying to identify which histone methyltransferases are involved by using knockout mice.”